Ymol
Ymol available freely for downloading!
Ymol is a molecular visualisation and animation program for the X Window System. It can be used to create production quality images, and movies, using either the built in renderer or by exporting the scenes to POVRAY (Persistence of Vision).
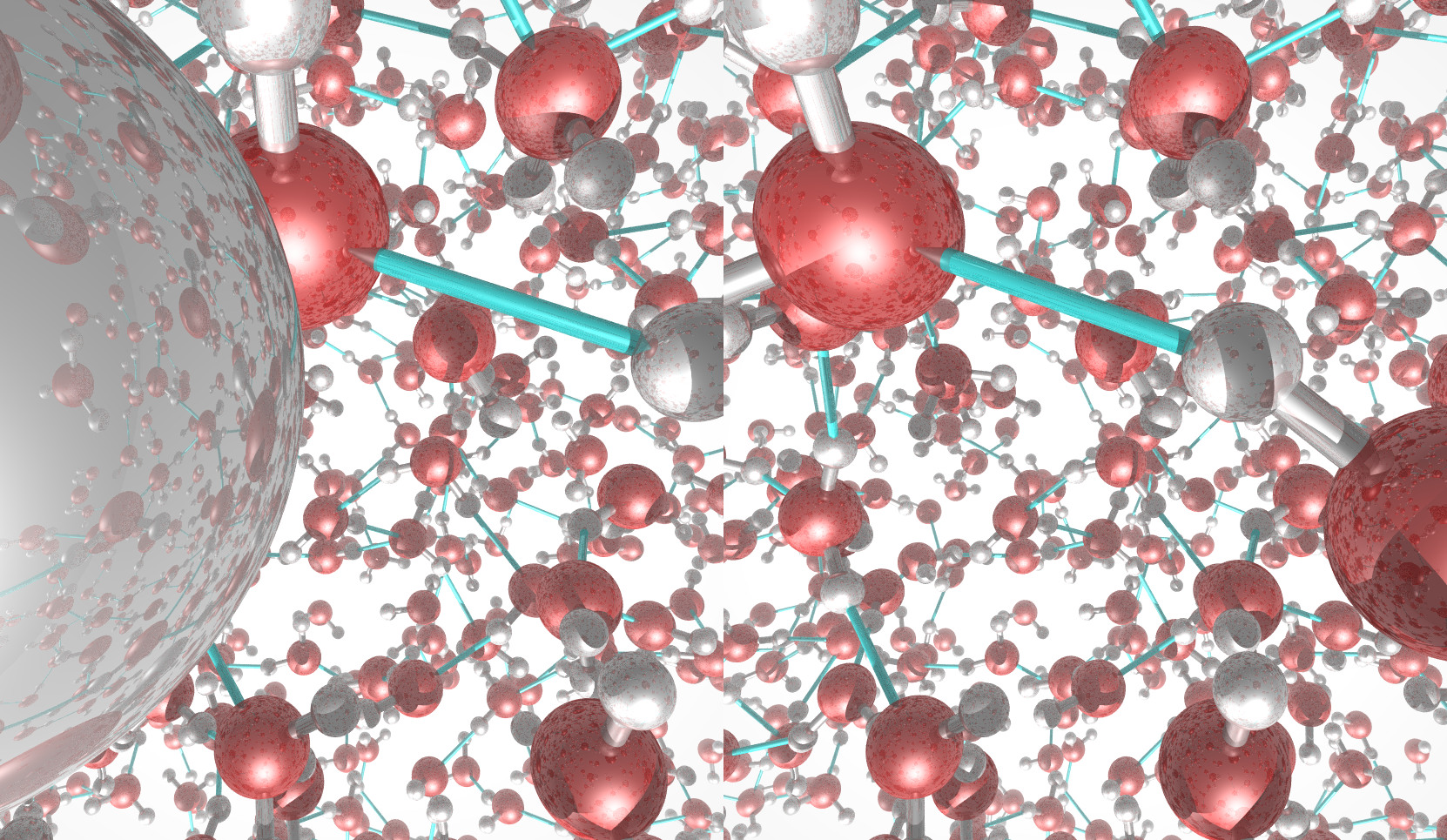
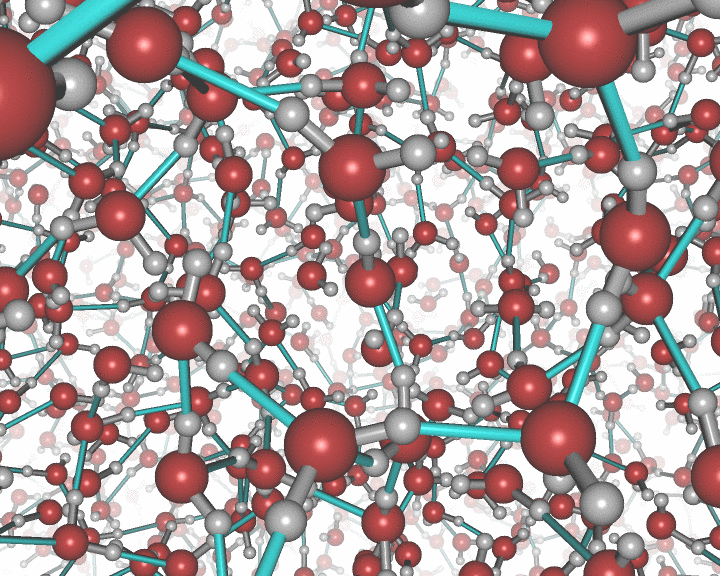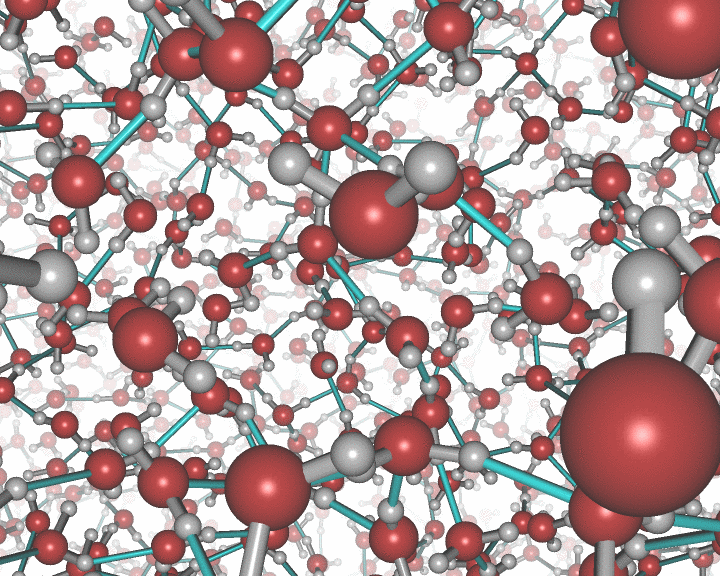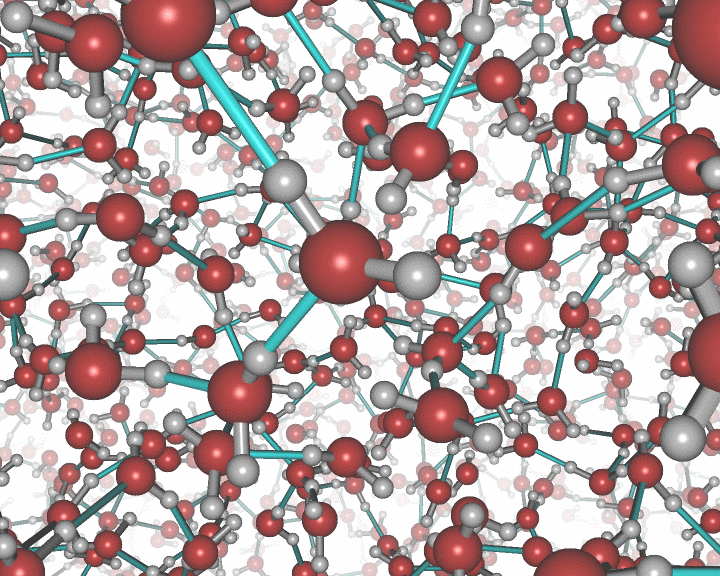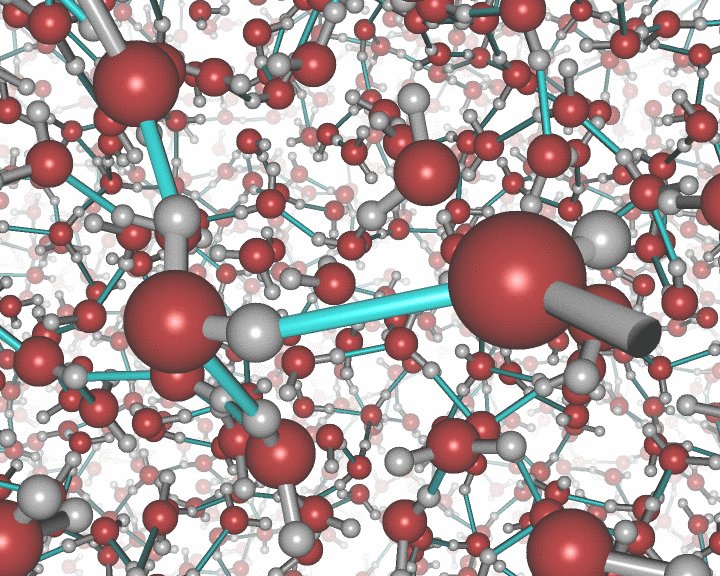
The rotating nanocluster below was rendered with Ymol.
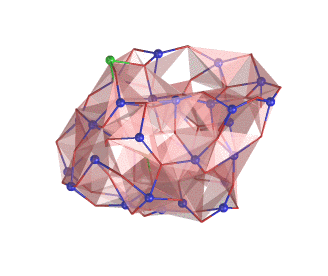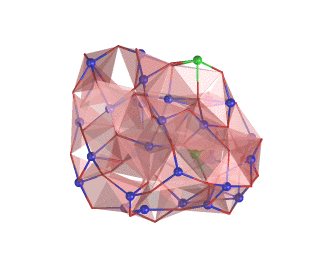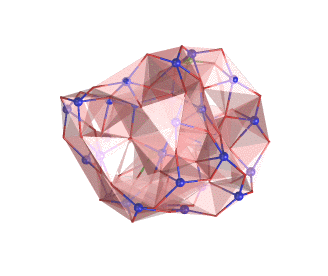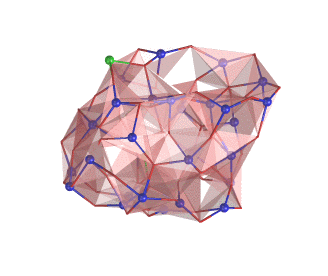
For more examples see the picture gallery.
The current version of ymol is 0.8.154. Ymol is available as a
binary and source distribution free for non commercial use. Although
development and testing mainly occurs on Linux (x86_64), Ymol has
been tested to run on many *nix-like operating system (Linux, OS X,
FreeBSD, IRIX, OSF/1, ULTRIX, AIX, Solaris, HP-UX (PA), and Win32
(Cygwin)). Ymol should be fairly easy to port to anything running
*nix and X11.
Pretty-looking pictures with many atoms might
take some time to render, but in the default mode Ymol operates very
fast and is very suitable for interactive use. Using only core Xlib
calls to draw circles, polygons and lines, Ymol shows real time
movies on slow (486) machines. Ymol can also use the POVRay raytracer.
Download ymol
Go to Ymol distribution directoryNOTE: New simpler installation from source
Only grab the ymol-allsource-VERSION.tar.gz archive. Unpack. Type make. Type make install. Done.Note though that this installs into ~/bin, so put this into your path:
PATH="$HOME/bin:$PATH"
export PATH
Latest updates
-
2018-08-29 (same day!)
Do not compute polygon rules when loading a style file if there are no polygon rules in the style file. Fix string length bug crash when saving orientation. Fix saving name of polygon rule. Released source and Linux (x86_64) binaries. Version 0.8.154.
-
2018-08-29
Two bugfixes related to polygon rules: Loading .ymol files with polygon rules sometimes made ymol crash when editing polygon rules. Reading files with predefined triangles did not work due to polygon rules always overwriting the triangles. Unfortunately predefined triangles and polygon rules cannot be used simultaneously, but at least now both can be used separately. Released source and Linux (x86_64) binaries. Version 0.8.153.
-
2018-07-03
Moved to new server. Made sure ymol still compiles with gcc 7.3.0. Just send a mail to daniel@spaangberg.se if you face any issues.
-
2014-07-25
Modified oversampling defaults. Increased buffer size (uses somewhat more memory). Moved to spaangberg.se. Released source and Linux (x86_64) binaries. Version 0.8.152.
-
2014-01-24
Fixed bug: When a style was imported the crystal cell setting was imported too, which should not happen. Released source and Linux (x86_64) binaries. Version 0.8.149.
2014-01-21
Fixed bug: Cells with non-zero off diagonal elements were displayed wrong with vaspymol (transposed H-matrix). Released source and Linux (x86_64) binaries. Version 0.8.148.
2013-12-18
Setting transparency=0 in polygon rules causes all rules for the central atoms and ligands to be applied, but the actual polygons are not drawn. Released source and Linux (x86_64) binaries. Version 0.8.147.
2013-12-18
Rearranged context sensitive menus (right button click). Released source and Linux (x86_64) binaries. Version 0.8.146.
2013-12-18
Added "Save style", and "Save global style". Added "Save orientation" and "Load orientation". Fixed selection menu bug. Fix so vaspymol can read also XDATCAR from vasp 5.2. Added more options to vaspymol: -1 expands the atoms but not the cell itself. Also added translation options. For instance "vaspymol -1 2 2 1 0.1 -0.1 0" will only expand the atoms 2 times in "x and y directions" (depending on cell vectors of course), no expansion in "z direction" and translate 0.1 -0.1 0 along the cell vectors. Released source and Linux (x86_64) binaries. Version 0.8.145.
2013-12-11
Fix so vaspymol can read several different versions of the XDATCAR format. Released source and Linux (x86_64) binaries. Version 0.8.144.
2013-12-10
Added vaspymol script (requires python). In your vasp directory run vaspymol to read XDATCAR, OUTCAR, POSCAR, CONTCAR files (XDATCAR is default unless it cannot be read in which case POSCAR will be read instead. Also POTCAR is needed to find atom types). The -p parameter chooses POSCAR, -o OUTCAR, -c CONTCAR. Additionally the cell can be expanded in the cell vector directions: "vaspymol 2 2 2" expands twice in all directions. Released source and Linux (x86_64) binaries. Version 0.8.143.
2013-09-23
Added polygon rules, allowing drawing of crystal structures. The polygon rules work similarly to the bond rules. Choose central atoms and ligands. Check the picture gallery for an example of this. Also made a simple crystal structure box translation code available. When a crystal structure has been read in (a box is needed of course!), translate it in the box by holding the shift button and using either cursor keys and PgUp/PgDown or the mouse motion or scroll wheel. Released source and Linux (x86_64, i386, ppc) binaries. Version 0.8.142.
2013-09-20
Updated defaults to use the "slice bond" option for covalent bonds, and new bonds. Released source and Linux (x86_64, i386, ppc) binaries. Version 0.8.141.
2013-09-19
Added possibility to have bonds where half the bond has the color of one atom and the other half the color of the other. See the "slice bond" option in bond properties and bond rules. The defaults stay the same as before. Check the picture gallery for an example of this. Released source and Linux (x86_64) binaries. Version 0.8.140.
2013-04-05
Made ymol Darwin target reflect Mac OS X mountain lion. Requires Xquartz installation before compilation. Released only the source due to this fix. Version 0.8.139.
2012-01-23
Fixed semi-transparant windows on compositing window managers, by more careful selection of visual. This also fixed a bug which drew all objects in black on some machines. Also added more window manager hints, and now finally name windows properly. Released source and Linux (x86_64) binaries for the 0.8.138 version.
2011-06-04
Made ymol easier to compile from source by providing a single archive, and a single make command to compile. Added explicit test for Darwin target (Mac OS X). Released source and Linux (x86_64) binaries for the 0.8.137 version.
2010-09-05
Add general support for case insensitive filesystems (Mac OS X). Released source and Linux (x86_64) binaries for the 0.8.136 version.
2010-08-20
Gaussian 98 import now reads Z-Matrix/Standard/Input orientation. Finally made gfortran the default in the source. Released source and Linux (x86_64) binaries for the 0.8.135 version.
2009-05-23
When deleting atoms all labels were removed. Fixed. Released source and Linux (i386, x86_64) binaries for the 0.8.134 version.
2009-03-30
Updated to fix sockimport from crashing if. Released source for the 0.8.133 version.
2007-10-22
Small change: default distance between eyes is 1 angstrom in stereo view. Version 0.8.132 (not publically released).
2007-10-17
Released source and Linux (i386, x86_64) binaries for the 0.8.131 version.
2007-10-17
Primarily many changes to the stereographic mode. It now works for all view modes, also with povray. See the image gallery for an example. Also a couple of rendering mode bug fixes.
2007-10-10
Released source and Linux (i386, x86_64) binaries for the 0.8.130 version.
2007-10-10
Added possibility to assign explicit bonds in the ymol socket interface. Check the testsockbond.for example.
2007-10-05
Released source and Linux (i386, x86_64) binaries for the 0.8.129 version.
2007-10-05
Added Triangelize mode.
2007-09-15
Released source and Linux (i386, x86_64) binaries for the 0.8.128 version.
2007-09-15
Wrote a ymol library which can be linked into other applications to ease the use of ymol as a realtime visualization package for other application. Wrote the ymolssh command to allow forwarding of the ymol ports used for visualization through a firewall.
2007-08-30
Released source for 0.8.127 version. Only difference is a removal of winsock test, which made ymol not compile under cygwin, so no need to release binaries for this version.
2007-08-12
Released source and Linux (i386, x86_64) binaries for the 0.8.126 version.
2007-08-12
Some minor fixes, such as cleanup of the ymolpid and ymol_sockimport files at program exit.
2007-08-10
Released source and Linux (i386, x86_64) binaries for the 0.8.125 version.
2007-08-10
Added new stereographic display mode.
2007-08-10
Released source and Linux (i386, x86_64) binaries for the 0.8.124 version.
2007-08-10
Added new socket interface to ymol. The testsocket.py python program in the ymol distribution demonstrates the use of this socket interface.
2007-02-14
Released source and Linux (i386, x86_64) binaries for the 0.8.123 version.
2006-11-02
Several bug fixes.
2006-10-13
Position and color of atomlabels can now be modified through the CLI. Several bug fixes.
2006-03-07
Released source, Linux (i386, x86_64), and FreeBSD (i386) binaries for the 0.8.120 version.
2006-03-07
Phong RSV and N can now be accessed easily through the default atom properties dialog.
2006-03-06
The default window size is now 720x576 to ease the creation of PAL video.
2006-03-06
Bondrules are now much faster to calculate, since a cell searching O(N) algorithm replaced the old O(N2).
2005-06-14
Can now read some files from the mdsus program.
2005-04-07
Released source for the 0.8.119 version.
2004-11-17
Can export data explorer from the CLI.
2004-05-28
User can load new fonts.
2004-05-22
Released source for the 0.8.118 version. Released source for Xco 0.2.136 and y4vm 0.8.22.
2004-05-12
Now supports x86_64.
2004-03-22
Released Linux (i386) binaries for the 0.8.118 version.
2003-11-24
Loop control widget added to control panel.
2003-11-24
The povray mode now honours the camera constant in the rendering parameters.
2003-05-01
Can now translate the molecules, not only zoom and rotate
2003-01-28
Released source for the 0.8.117 version.
Picture gallery
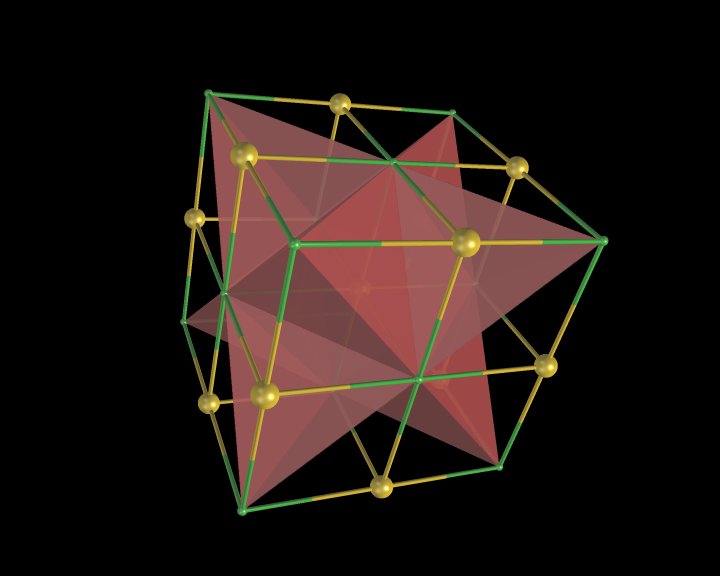 An example of the new polygon rules: NaCl crystal box.
An example of the new polygon rules: NaCl crystal box.
-
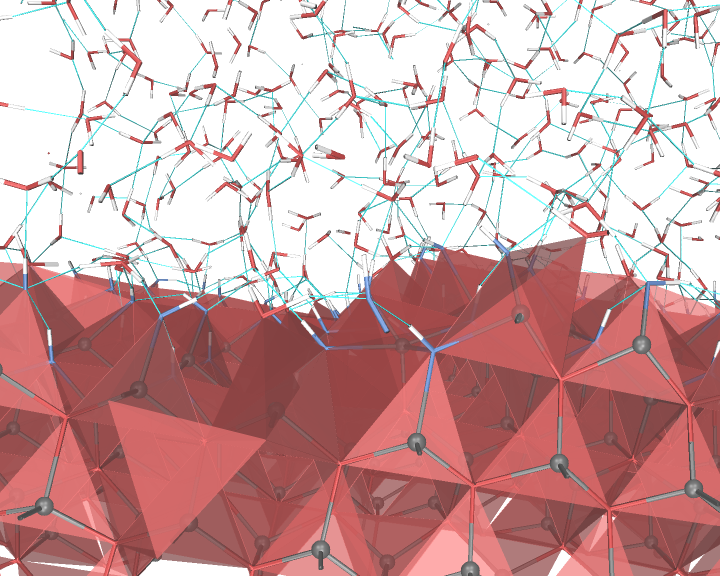 An example of the new polygon rules: ZnO/water interface.
An example of the new polygon rules: ZnO/water interface.
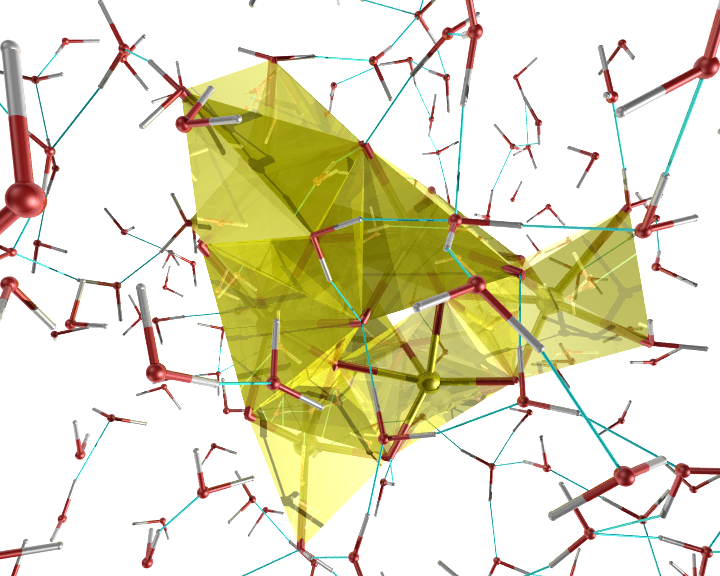 An example of the new polygon rules: A nanocluster in liquid.
An example of the new polygon rules: A nanocluster in liquid.
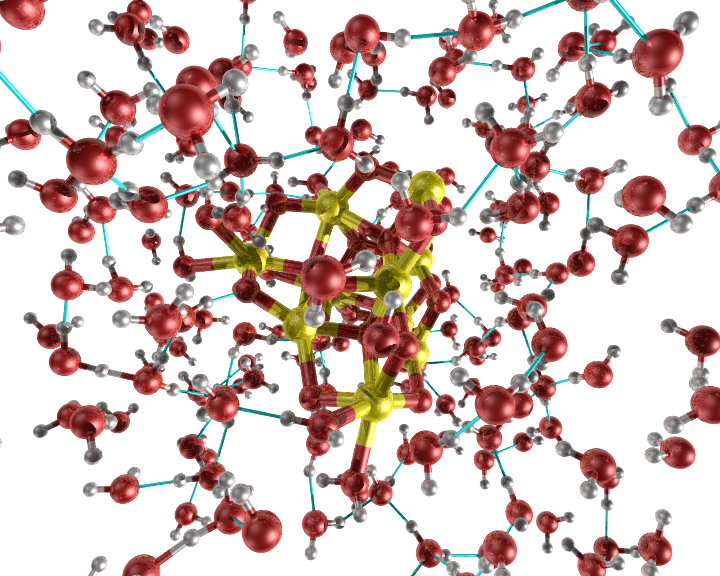 An example of the new bond option "slice bond".
An example of the new bond option "slice bond".
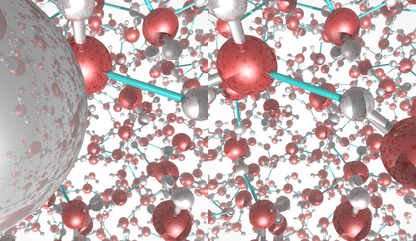 This picture was generated with the stereo mode and the povray
raytracer. The image portrays liquid water.
This picture was generated with the stereo mode and the povray
raytracer. The image portrays liquid water.
 An animation of liquid water (a big
gif animation!).
An animation of liquid water (a big
gif animation!).
The ymol library
The ymol library makes it easy to use ymol as a visualization package. An example C-program:
#include <stdio.h>
#include <math.h>
#include "ymolsock.h"
int main()
{
double t=0.;
int n=200;
double radius=5.;
double step=1./(n*50);
double pi=3.1415926;
ymolsock_init();
while (1)
{
int i;
if (ymolsock_check_ymol())
{
if (ymolsock_write_natoms(n))
{
for (i=0; i<n; i++)
{
double x=radius*sin(3*(t+i*pi*2/n));
double y=radius*cos(5*(t+i*pi*2/n));
double z=radius*cos(7*(t+i*pi*2/n));
ymolsock_write_atom(i,1,x,y,z);
}
}
}
t+=pi*2*step;
}
ymolsock_deinit();
return 0;
}
An example Fortran-program:
program testsock
implicit double precision (a-h,o-z)
t=0.d0
n=200
r=5.d0
step=1.d0/(n*50);
pi=3.1415926
call yscini
10 call yscchy(ierr)
if (ierr.ne.0) then
call yscwna(n,ierr)
if (ierr.ne.0) then
do i=1,n
x=r*sin(3*(t+i*pi*2/n))
y=r*cos(5*(t+i*pi*2/n))
z=r*cos(7*(t+i*pi*2/n))
call yscwat(i,1,x,y,z,ierr)
enddo
endif
endif
t=t+pi*2*step
goto 10
call yscdin
stop
end
When ymol is compiled and installed as an ordinary user on a Linux
x86_64 platform an include file and a library is installed in
$HOME/lib/Linux.x86_64. The C program above can then easily be
compiled using:
gcc -O2 -o testsock testsock.c -L$HOME/lib/Linux.x86_64 -I$HOME/lib/Linux.x86_64 -lymol -lmThe Fortran program above is easily compiled as well using:
g77 -O2 -o testsock testsock.for -L$HOME/lib/Linux.x86_64 -lymol
The ymolssh command
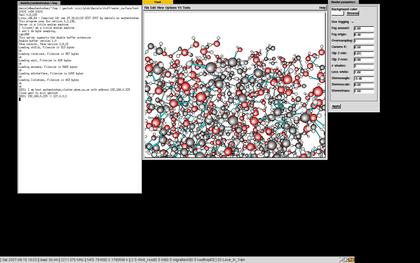
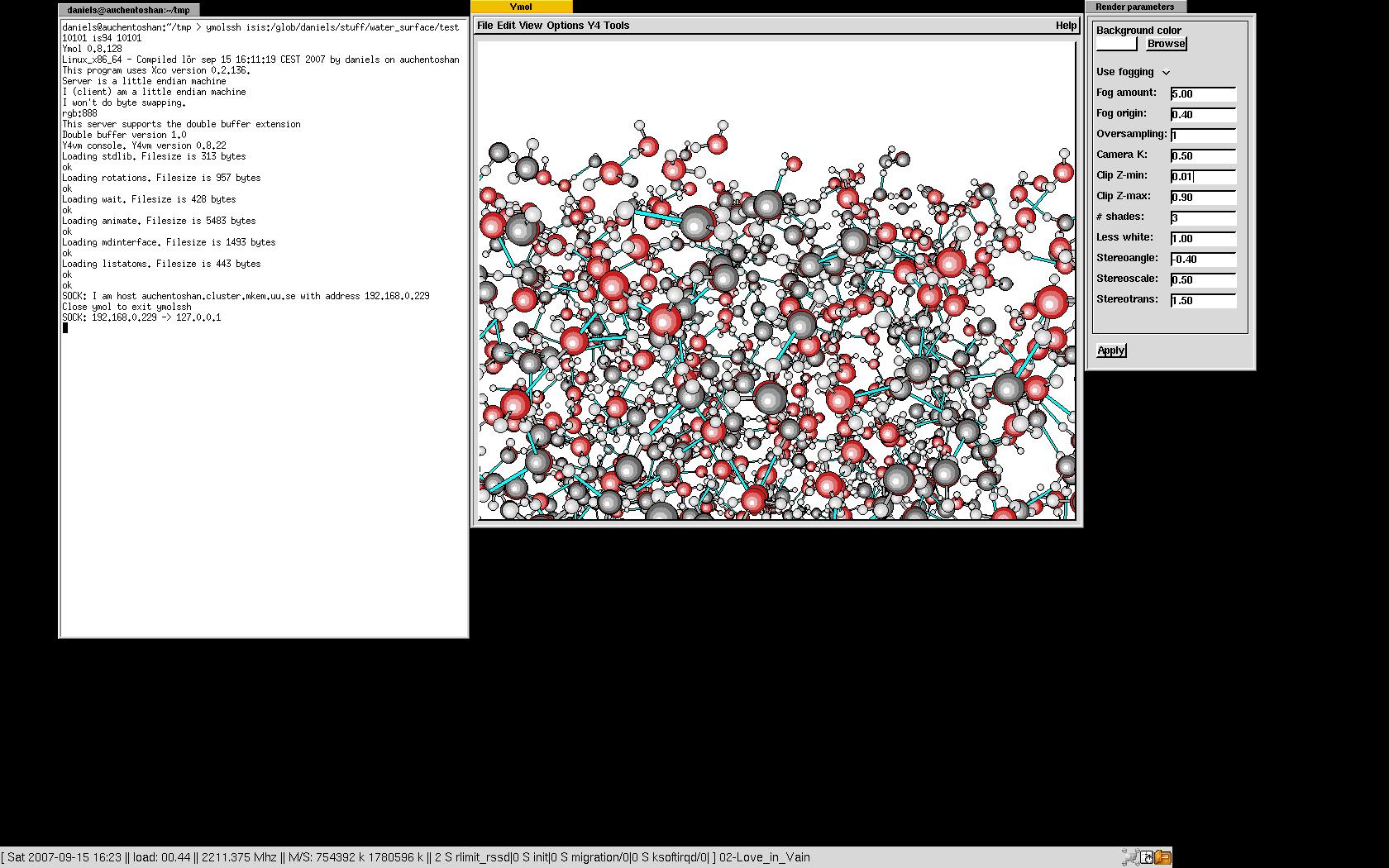
The ymolssh command can be used to visualize a simulation running on a remote computer if you or the remote computer is behind a firewall. If there's no firewall in between, it is only necessary to start ymol and copy the "ymol_sockimport" to the directory where the simulation is running. If the simulation is running on the computer named simmachine in the directory simdirectory it is only necessary to type
ymolssh simmachine:simdirectory 10201The port (10201) on the remote host (simmachine) used to forward the ymol data is necessary to give as well. Use any unused port. Should the simulation be running on a remote computer with no ssh-access, it may be necessary to go to that machine in steps. This is also supported by ymolssh:
ymolssh simmachinefrontend:simdirectory 10201 simmachine 10201See the image for a real example of this.